A couple of weeks ago the Biochemistry Graduate Association Gyanu Lamichhane to give a seminar on his work on peptidoglycan transpeptidases. In his very nice talk Lamichane told the story of how a search for transposon mutants with virulence phenotypes led him to work on an unexpected peptidoglycan transpeptidase. Lamichhane’s talk prompted me to look at what is needed to represent peptidoglycan transpeptidases in the Gene Ontology. This post is my notes so far on figuring out what is needed.
Some background is in order to explain why this enzyme was unexpected. Peptidoglycan makes up the bacterial cell wall. Because it’s not found in eukaryotes (e.g. humans), and it’s needed to keep bacterial cells from exploding, peptidoglycan synthesis has long been the target of clinically important antibiotics. In the textbook version of peptidoglycan, an intermediate precursor is constructed where a pentapeptide is built up on a UDP-MurNac. This MurNac-pentapeptide is linked to a UDP-GlcNac to make a disaccharide with an attached pentapeptide. This is transferred to a carrier, flipped to the outer surface of the inner membrane, and polymerized into the growing peptidoglycan. Crosslinks catalyzed by peptidoglycan transpeptidases create a polypeptide orthogonal to the polysaccharides.
In many organisms, including E. coli and M. tuberculosis, the unit can be written as:
GlcNac-MurNac-(L-Ala1-D-iso-Glu2-m-DAP3-D-Ala4-D-Ala5)
in others, including Enterococci, it’s
GlcNac-MurNac-(L-Ala1-D-iso-Glu2-L-Lys3-D-Ala4-D-Ala5)
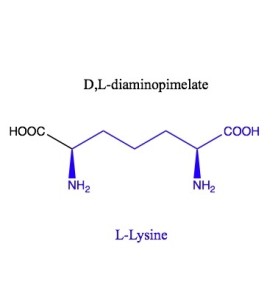 m-DAP is meso-Diaminopimelic acid, also known as D, L-Diaminopimelic acid is made in a few different ways, but is an intermediate in both peptidoglycan and lysine biosynthesis. Lysine is made by decarboxylating the D end of mDAP. The L end is what gets incorporated into peptidoglycan.
m-DAP is meso-Diaminopimelic acid, also known as D, L-Diaminopimelic acid is made in a few different ways, but is an intermediate in both peptidoglycan and lysine biosynthesis. Lysine is made by decarboxylating the D end of mDAP. The L end is what gets incorporated into peptidoglycan.
The textbook version of peptidoglycan has crosslinks made by a transpeptidation reaction where the D end of mDAP (the acceptor) replaces the terminal D-ala on another pentapeptide. This could be called a 4-3 D,D transpeptidation reaction with the D-Ala from the 5 position being the leaving group.
Lamichhane’s transpeptidase links two m-DAPs at the 3 positions directly together, with the side chain of one attacking the backbone of the other, with the release of D-Ala-D-Ala. The donor is the L end of the mDAP, so I think this is why I’d call it a 3-3 L,D transpeptidation. The nomenclature here is related to, but not identical to, what I’m seeing in the papers, where they talk about 4-3 and 3-3 crosslinks and D,D and L,D transpeptidases.
The reason we need names that are more specific than L,D and D, D is that it gets more complicated. In E. coli, in addition to the D,D 4-3 transpeptidases there are five L,D transpeptidases. Two make direct 3,3 links like Lamichane’s enzyme from TB. Three others use the ε NH2 of the C-terminal Lysine of Lpp (aka Braun’s lipoprotein) as the acceptor, resulting in attachment of Lpp to the peptidoglycan.
Meanwhile, in Enterococcus, what I wrote is oversimplified L-Lys replaces m-DAP at the 3 position, but sometimes the Lysine is modified to form things like (N ε-D-Asx)-L-Lys in E. faecium. And this just scratches the surface of chemical diversity in bacterial peptidoglycan.
[pmid-refs key=’peptidoglycan transpeptidases’ limit=10]